The MIF is located within PC2 laboratories, allowing analysis of both Class 2 pathogens and genetically modified organisms along with cell culture and pathogen preparation rooms.
The MIF houses a specialised imaging system for structured illumination microscopy that is unique in NSW. This system can image cells in real-time at high resolutions. As the first adopter of this technology in Australia MIF has developed special expertise in imaging living microbes using this system.
Microbial imaging at the MIF has led to ground-breaking insights published in high quality journals such as Nature, Nature Microbiology, and Nature Communications. Selected examples of how research enabled by MIF has changed the way we think about bacterial behaviour include:
Along with this, MIF offers routine optical microscopy techniques such as laser-scanning confocal microscopy, wide-field fluorescence microscopy, phase-contrast/DIC microscopy, live-cell imaging and high-content imaging.
MIF also provides advanced image analysis capabilities and expert advice. Along with this they offer computational labs, software and access to UTS’s Data Arena, a 360-degree interactive data visualisation facility.
Microorganisms such as protozoa and bacteria have been conducting an “arms race” for billions of years. Researchers at UTS, as part of an international team, have discovered that for pathogenic bacteria, this arms race may have made them not only stronger but much more infectious. The results provide a new understanding about the mechanisms of infection and disease transmission.
This discovery could help explain why the cholera-causing bacteria V. cholerae, is so persistent in aquatic environments. The researchers observed that strains of V. cholerae expelled into the environment after ingestion by protozoa were primed for colonisation and infection in humans. After protozoa consume their food, it is encased in a membrane. Cholera bacteria are also encapsulated in this membrane protecting them both inside the protozoa and after they are ejected into the environment. These capsules are called ‘expelled food vacuoles’ (EFV). The results show that pathogenic bacteria encased in EFVs are resistant to starvation, antibiotics and the acidic conditions encountered in the human stomach.
Confocal microscopy at the (MIF) was used to study the escape of V. cholerae from the EVFs and confirm the presence, number and viability of EFVs in cultures. Transmission electron microscopy was also performed at another of our facilities – the Electron Microscope Unit at UNSW – to observe the relationship between the protozoa and the V. cholerae encapsulated within the EFVs.
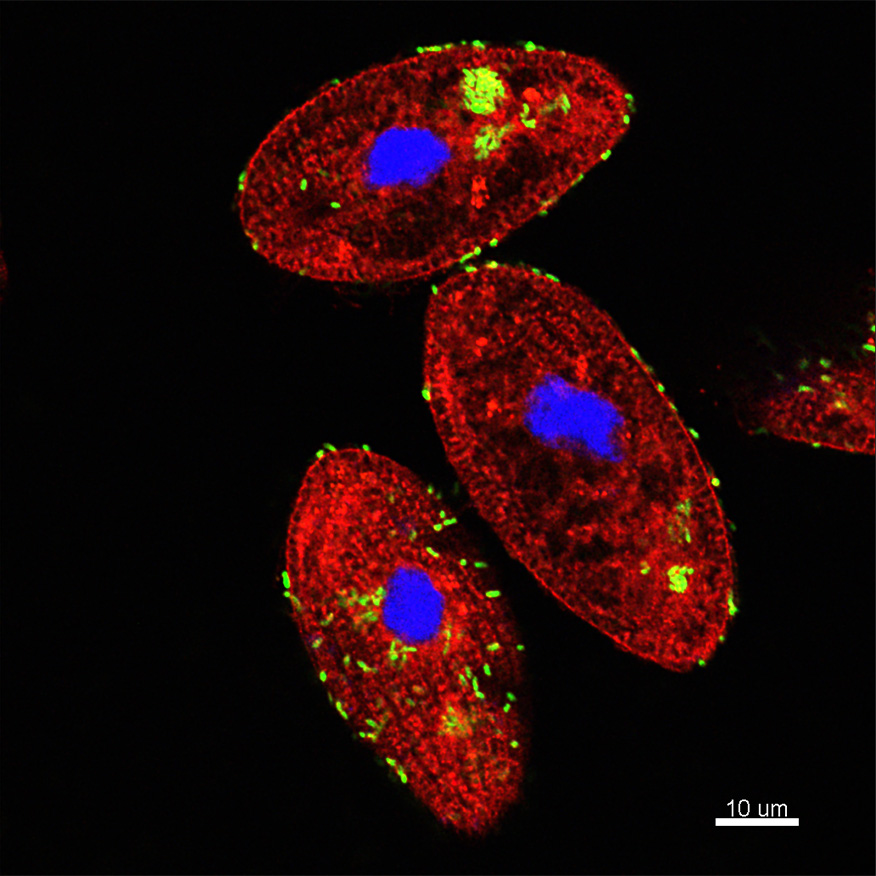
The micrograph shows Tetrahymena pyriformis (aquatic protozoan ciliate) co-incubated with Vibrio cholerae under natural conditions. Co-incubation results in the ingestion of V. cholerae by T. pyriformis into phagosomes or food vacuoles. Green bacteria can be observed inside phagosomes and also outside around the cell. The intracellular bacteria remain packaged and viable inside the food vacuoles and then are released as egested food vacuoles (EFVs) to the extracellular space, protecting V. cholerae from environmental stressors and significantly enhancing its infectious potential. Red dye: FM4-64FX which stained phospholipids, Green: V. cholerae-GFP, Blue: DAPI staining the protozoan nucleus.
“Our hypothesis is that these bacteria have adaptive traits that have allowed them to either escape or survive being eaten by protozoan predators. These traits have evolved in the environment as a response to predation and not for the ability to cause disease in accidental hosts. Unfortunately, these adaptive traits can increase the ability of the bacteria to colonise humans and cause disease,” A/Prof. McDougald said.
Future research could identify biomarkers, such as proteins on the surfaces of EFVs, so antibodies could be developed to enable real-time monitoring of outbreaks.
“The discovery of this highly infectious form of cholera produced outside the human body provides an opportunity to detect it and reduce the incidence of cholera. This disease remains a global threat to public health, particularly in developing countries,” Prof Harry, former director of the ithree institute at UTS, said.
Aquatic protozoan ciliate incubated with cholera – see case study for more details.
September 27, 2020